Fast nonnegative spatiotemporal calcium smoothing in dendritic
trees
Eftychios A. Pnevmatikakis*, Columbia University, eftychios@stat.columbia.edu
Keith Kelleher*, University of Houston, keith.kelleher@gmail.com
Rebecca Chen, University of Houston, rlchen83@gmail.com
Kresimir Josić, University of Houston, josic@math.uh.edu
Peter Saggau, Baylor College of Medicine, psaggau@bcm.tmc.edu
Liam Paninski, Columbia University, liam@stat.columbia.edu
* Equal contributions
Submitted to COSYNE11
Summary
Understanding what triggers synaptic strength modifications in vivo
remains a key problem in cellular neuroscience. Recent fast scanning
multi-photon microscopy techniques [1] support the role of calcium as
a key biochemical effector, signaling the coincident occurrence of
back-propagating action potentials (bAPs) and excitatory post-synaptic
potentials (EPSPs), which is a key tenet of models of STDP
[2]. However, determining the entire spatio-temporal pattern of
calcium influx is difficult since the available experimental
techniques are noisy, sub-sampled observations of the true underlying
calcium signals. It is therefore necessary to use statistical methods
to infer details of calcium transients. Optimal spatiotemporal
smoothing of the calcium profile on a dendritic tree given local noisy
measurements remains a computationally hard problem, due to the high
dimensionality and complex structure of dendritic trees.
Here we take a functional approach: The evolution of calcium
concentration on the whole tree is determined from a smaller set of
hidden variables that govern the calcium dynamics and incorporate
possible calcium bumps due to bAPs, EPSPs or external stimulation. The
observations are then expressed as linear, noisy measurements of the
hidden variables. Using a state-space approach, our problem reduces to
the maximum a posteriori estimation of these hidden states and can be
efficiently solved, if the prior distributions of the calcium activity
and measurement noise are log-concave as a function of the hidden
states. The complexity of the estimation algorithm scales linearly
both with the number of time steps T over which we infer the
underlying signal and with the number of hidden states d (i.e., the
size of the tree), leading to a tractable overall complexity of
O(dT). We apply our algorithm to real reconstructed dendritic trees
and find that the filtered output can quite accurately interpolate and
denoise the subsampled, noisy observed spatiotemporal data.
Here is an example:
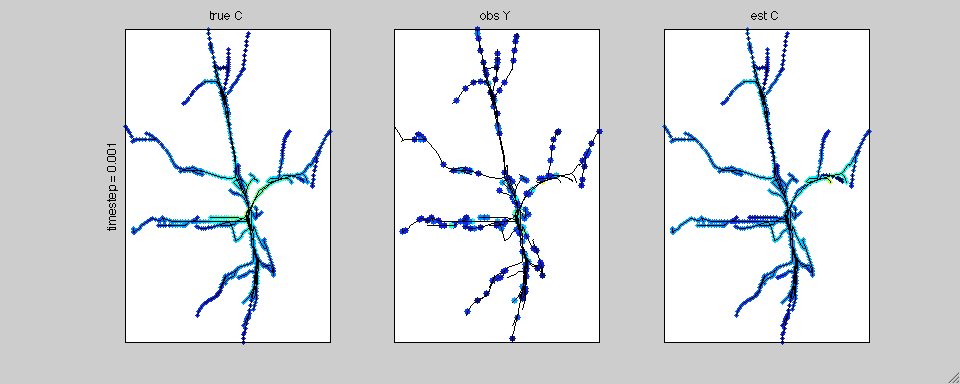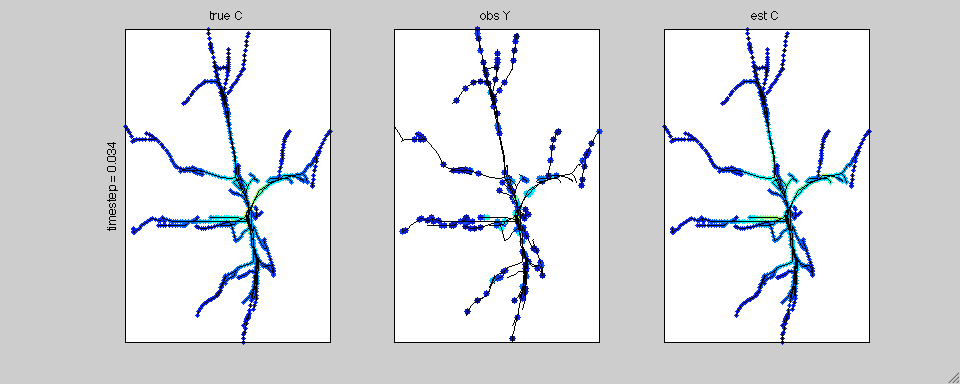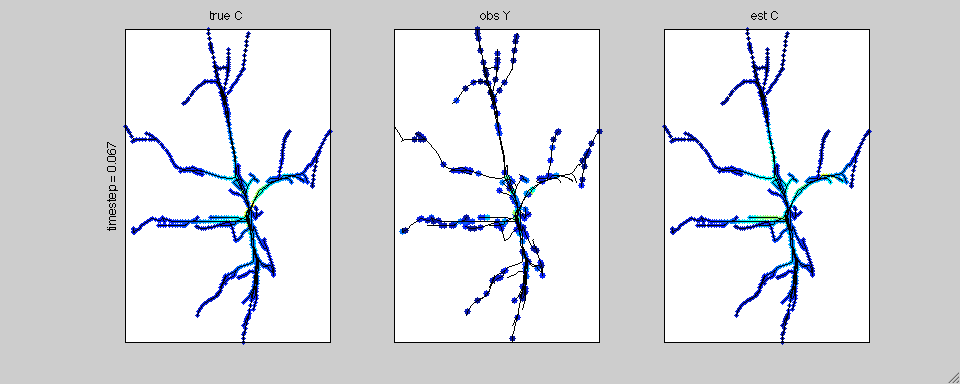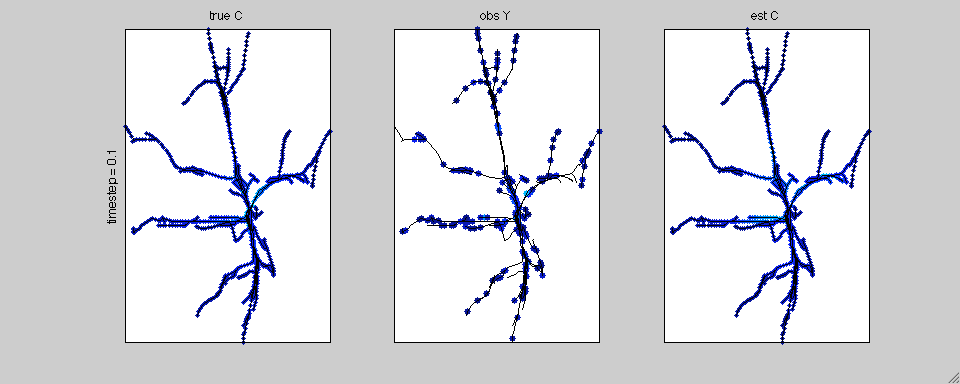
In this case we have simulated calcium transients on the reconstructed
anatomy of a hippocampal pyramidal cell. The true calcium data is
shown on the left; the noisy, subsampled observations are shown in the
middle; and the estimated spatiotemporal calcium profile is shown on
the right.
Methods
The calcium activation is modeled as a linear combination of localized
spatial functions (e.g. splines or diffusion kernels) weighted by
hidden, temporal variables. As a result, the calcium profile on the
whole tree is given by the multiplication of the hidden variables with
a sparse tree-banded matrix. The temporal dynamics are modeled by an
auto-regressive process driven by non-negative inputs that model
calcium bumps. The distribution over the inputs models our prior
knowledge about calcium influx and local correlations, and can be
chosen to be log-concave in general. In our data the measurements,
taken from a fast three-dimensional scanning microscope, have the form
of emitted light at certain positions and are corrupted with highly
non- gaussian, yet log-concave, noise. The MAP estimate of the hidden
variables is the solution of an optimization problem, which due to
log-concavity can be found efficiently using standard interior-point
algorithms. Our model is of first order hidden markov form, and
therefore the Hessian of the log-posterior is a block tridiagonal
matrix which can be inverted in O(d3T) time and space using a
forward-backward recursion (e.g. the LDL decomposition). As the
number of compartments can often be quite large, it is important to
seek methods with complexity that does not scale superlinearly with
d. By exploiting the tree-banded structure of the problem, we find
that each matrix in the LDL recursion is sparse and approximately
tree-banded. Consequently, the complexity and space requirement can be
further reduced to O(dT), and therefore applied to arbitrarily complex
dendritic structures and arbitrarily long experiments.
References
[1] Reddy GD et al, Nature neuroscience, 11(6):713-720, 2008.
[2] Sjostrom PJ, Nelson SB, Current opinion in neurobiology, 12(3):305-314, 2002.

